Synthetic Data
Part of the MapEHR is a data synthesizer for generating clinically relevant data instances for openEHR.
The MapEHR engine uses operational templates (OPT2) and YAML based rules to specify clinically relevant data for generating openEHR compositions for storage in an openEHR repository. These YAML rules are just another source for the engine.
Introduction
The Basics
Rules are stored in YAML files. Data synthesizer receives a list of directories in which it searches for the rules. The directory structure is up to the users – synthesizer searches all sub-folders of the received directories.
Each YAML file may contain one or more rules (under rules:).
Rules may be specified for: loinc, snomed, archetypes, templates.
Example for loinc:
rules:
# LOINC based formulas
loinc:
# Heart rate
8867-4:
Example for archetypes:
rules:
# Archetype based formulas
archetypes:
# Heart rate
openEHR-EHR-OBSERVATION.t_vital_signs-heart_rate.v1:
Set a value for ELEMENT.value
A simple formula to set a ELEMENT.value attribute for a LOINC code:
# Formulas
rules:
# LOINC based formulas
loinc:
# Heart rate
8867-4:
uri: http://loinc.org/8867-4
name: Heart rate
set:
- attribute: value
element:
value_interval: 40..130
In the above example 8867-4 is the LOINC code for heart rate. It is found in the openEHR-EHR-COMPOSITION.t_encounter-vital_signs.v1.0.0.opt2 for the id0.2 (archetype openEHR-EHR-OBSERVATION.t_vital_signs-heart_rate.v1.0.0):
["id5"] = <http://loinc.org/8867-4>
The set: contains a single "set" instruction for attribute: value. The value in this case refers to ELEMENT.value since the LOINC code 8867-4 will find an ELEMENT element.
The element: uses value_interval: 40..130 to inform the data synthesizer to pick a random value in the interval 40..130 and set it to ELEMENT.value element. In this case it is DV_QUANTITY element so the random value will be set to ELEMENT.value.magnitude. Units were already set so we don't need to change them.
The interval in value_interval: will be used to set ELEMENT.value.normal_range.
Set a value for ELEMENT.value - multiple units
Another simple formula to set a ELEMENT.value attribute for a LOINC code, but this time the value may be in different units:
# Formulas
rules:
# LOINC based formulas
loinc:
# Body temperature
8310-5:
uri: http://loinc.org/8310-5
name: Body temperature
set:
- attribute: value
element:
value_intervals:
deg_C_snomed: 35.0..38.9
deg_F_snomed: 90.0..102.0
interpretation_intervals:
deg_C_snomed:
low: 36.1
high: 38.0
deg_F_snomed:
low: 96.98
high: 100.4
In the above example 8310-5 is the LOINC code for body temperature. It is found in the openEHR-EHR-COMPOSITION.t_encounter-vital_signs.v1.0.0.opt2 for the id0.7 (archetype openEHR-EHR-OBSERVATION.body_temperature.v2.0.0):
["id5"] = <http://loinc.org/8310-5>
The set: contains a single "set" instruction for attribute: value. The value in this case refers to ELEMENT.value since the LOINC code 8310-5 will find an ELEMENT element.
The element: uses value_intervals: (notice plural) to inform the data synthesizer to pick a random value in the intervals based on the units found in the element (in this case ELEMENT.value.units). deg_C_snomed and deg_F_snomed are defined in the data-synth-formulas/formulas/units.map.yaml since they are used in multiple formulas.
In this case the ELEMENT.value element is DV_QUANTITY element so the random value will be set to ELEMENT.value.magnitude. Units were already set so we don't need to change them.
The interval in value_intervals: will be used to set ELEMENT.value.normal_range.
In addition the element specifies interpretation_intervals: which will be used to set ELEMENT.value.interpretation (not yet part of the openEHR RM).
Use choices
An elements value may use a list of choices from which a random item is picked:
rules:
loinc:
74720-4:
uri: http://loinc.org/74720-4
name: Device name
set:
- attribute: value
element:
value_type: DV_CODED_TEXT
choices:
- code: http://snomed.info/id/309641003
description: Aneroid sphygmomanometer
- code: http://snomed.info/id/466093008
description: Automatic-inflation electronic sphygmomanometer, non-portable
- code: http://snomed.info/id/466086009
description: Automatic-inflation electronic sphygmomanometer, portable, arm/wrist
In the above example the ELEMENT.value is a DV_CODED_TEXT. A random item in the choices: list will be selected and used to populate the DV_CODED_TEXT element:
"value": {
"_type": "DV_CODED_TEXT",
"value": "Aneroid sphygmomanometer",
"defining_code": {
"_type": "CODE_PHRASE",
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "snomed",
},
"code_string": "309641003",
}
}
Use choices to remove elements
A list of choices from which a random item is picked can also remove elements if they are not required:
rules:
loinc:
# Problem diagnosis
75326-9:
uri: http://loinc.org/75326-9
name: Problem
set:
- attribute: data
elements:
diagnosis:
value_type: DV_CODED_TEXT
body_site:
value_type: DV_CODED_TEXT
variant:
value_type: DV_CODED_TEXT
define:
diagnosis: http://loinc.org/29548-5
body_site: http://loinc.org/39111-0
variant: http://loinc.org/22689c4b-eb6e-ef11-8870-6045bdc71c8b
choices:
- diagnosis:
code: http://snomed.info/id/73211009
description: Diabetes mellitus
body_site:
code: http://snomed.info/id/278198007
description: Entire cardiovascular system
# If the [variant] is present, it will remain the data instance.
variant:
code: http://snomed.info/id/?
description: ???
- diagnosis:
code: http://snomed.info/id/193028008
description: Sick headache
body_site:
code: http://snomed.info/id/302548004
description: Entire head
# If the [variant] is not present, it will be removed from the data instance.
# variant:
In the above example the variant for the 1st choice will remain in the data instance. For the 2nd choice the variant element will be removed.
Use fake data
Formulas can use a "faker" library to set values.
rules:
loinc:
# Clinical interpretation
64110-0:
uri: http://loinc.org/64110-0
name: Clinical interpretation
set:
- attribute: value
element:
value: faker.loremParagraph()
value_type: DV_TEXT
In the above example the value: will be set to a random paragraph text.
Set RM attributes
Formulas can set multiple RM attributes in a set: instruction.
rules:
loinc:
79191-3:
uri: http://loinc.org/79191-3
name: Patient demographics panel
set:
- attributes:
source:
attributes:
id:
value: faker.datatypeUuid()
type:
value: faker.loremWord()
target:
attributes:
id:
value: faker.datatypeUuid()
type:
value: faker.loremWord()
The LOINC code 79191-3 will find an element of type Party_relationship (not part of openEHR RM). In the above example the attributes: refers to Party_relationship RM type. In this case we want to set attribute id and type inside the Entity_relationship.source and Entity_relationship.target attributes.
Set multiple elements
Formulas can set multiple elements for a set: instruction.
rules:
loinc:
90055-5:
uri: http://loinc.org/90055-5
name: Organization information panel
set:
- attribute: items
elements:
name:
value: faker.companyName()
value_type: DV_TEXT
national_provider_id:
value: faker.datatypeUuid()
value_type: DV_TEXT
role:
value: faker.personJobSector()
value_type: DV_TEXT
define:
name: http://loinc.org/76469-6
national_provider_id: http://loinc.org/76468-8
role: http://loinc.org/104974-1
In the above example the attribute: items refers to CLUSTER.items which is a list of elements. In this case all the elements we will set inside the CLUSTER.items are of type DV_TEXT. We use a generic "faker" library to set the DV_TEXT.value attribute.
The LOINC code 90055-5 will find an archetype openEHR-EHR-CLUSTER.person.v1.0.0 which is of type CLUSTER. Data synthesizer will continue to search for elements inside this CLUSTER.items. The elements are specified with elements: (notice plural). Each element uses a key which is fully defined in the define:.
For example to set the name element, the data synthesizer will search the CLUSTER.items for the LOINC code 76469-6. If found, its ELEMENT.value.value will be set to a random company name using the faker library expression: faker.companyName().
Use other elements to calculate a value
Sometimes we need to use other elements to calculate a value of another element. Calculating Body Mass Index (BMI) is such an example.
We use LOINC code 85353-1 to find the nearest common ancestor of all the elements we need. In this case the LOINC code is specified for a COMPOSITION (see openEHR-EHR-COMPOSITION.t_encounter-vital_signs.v1.0.0.opt2).
We use attribute: content to select COMPOSITION.content which is a list of OBSERVATIONs. For the BMI we need weight and height.
We use weight_observation element (with LOINC code 29463-7) to first select the OBSERVATION for openEHR-EHR-OBSERVATION.t_vital_signs-weight.v1.0.0. We need to go one level deeper to find the actual value for weight. This is achieved with using attribute: data to search inside OBSERVATION.data list. We use weight element (with LOINC code 29463-7) to find the weight CLUSTER. Note that in this case the OBSERVATION and the CLUSTER we need, both use the same LOINC code. This is why we defined weight_observation and weight separately even if they use the same LOINC code. The difference is that the first one holds the OBSERVATION element and the second one the CLUSTER element. We will use the weight of type CLUSTER in the bmi element.
Similarly we use height_observation and height to read the value for the height.
The BMI is in the OBSERVATION for the LOINC code 59574-4 and inside its OBSERVATION.data is an ELEMENT for the LOINC code 59574-4 (note the same LOINC code is used here too).
The bmi element uses a bmi() function to calculate its value:
value: bmi($weight, $height, $default_bmi)
rules:
loinc:
85353-1:
uri: http://loinc.org/85353-1
name: Vital signs, weight, height, head circumference, oxygen saturation and BMI panel
set:
- attribute: content
elements:
weight_observation:
attribute: data
elements:
weight: # We only need the existing value for the bmi().
height_observation:
attribute: data
elements:
height: # We only need the existing value for the bmi().
bmi_observation:
attribute: data
elements:
bmi:
value: bmi($weight, $height, $default_bmi)
interpretation_interval:
low: 18.5
high: 24.9
vars:
# Normal Distribution values for BMI:
# Source: https://www.scirp.org/journal/paperinformation?paperid=117728
# Default mean in randomNormalDistribution() is an average of male and female means.
default_bmi: randomNormalDistribution((27.6863+25.4960)/2, sqrt(18.65))
define:
weight_observation: http://loinc.org/29463-7 # Same LOINC code is used for OBSERVATION and CLUSTER.
weight: http://loinc.org/29463-7
height_observation: http://loinc.org/8302-2 # Same LOINC code is used for OBSERVATION and CLUSTER.
height: http://loinc.org/8302-2
bmi_observation: http://loinc.org/59574-4
bmi: http://loinc.org/59574-4
Examples of using the bmi() function:
-
Without defaults:
value: bmi($weight, $height) -
Use variance directly in randomNormalDistribution():
value: 'bmi($weight, $height, randomNormalDistribution((27.6863+25.4960)/2, (4.4351+4.2031)/2))' -
Using (unnecessary) long way to specify an expression:
value: |
var weight = $weight;
var height = $height;
if (weight == null || height == null) {
return randomNormalDistribution((27.6863+25.4960)/2, sqrt(18.65));
} else {
return bmi(weight, height);
}
How to add autocomplete for the rules in IntelliJ IDE
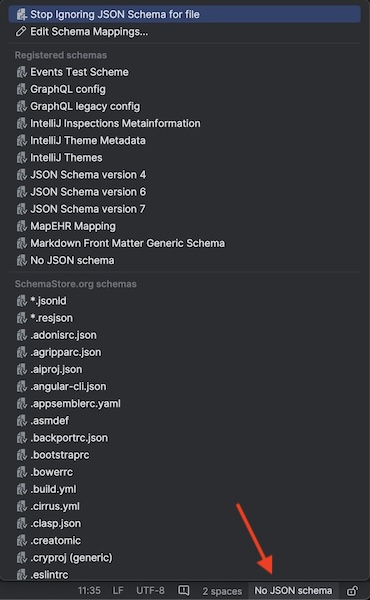
- Open one of the rule files (with
.map.yamlextension). - In the lower right click on
No JSON schema:
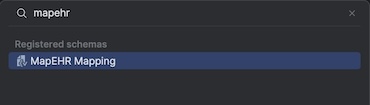
- Start typing
mapehrand selectMapEHR Mapping:
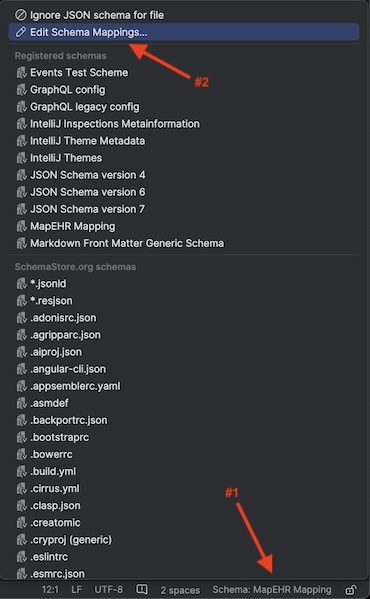
- In the lower right click on
Schema: MapEHR Mappingand selectEdit Schema Mappings...:
-
Under the
Schema version:click on the plus (+) icon and addFile path pattern: *.map.yaml: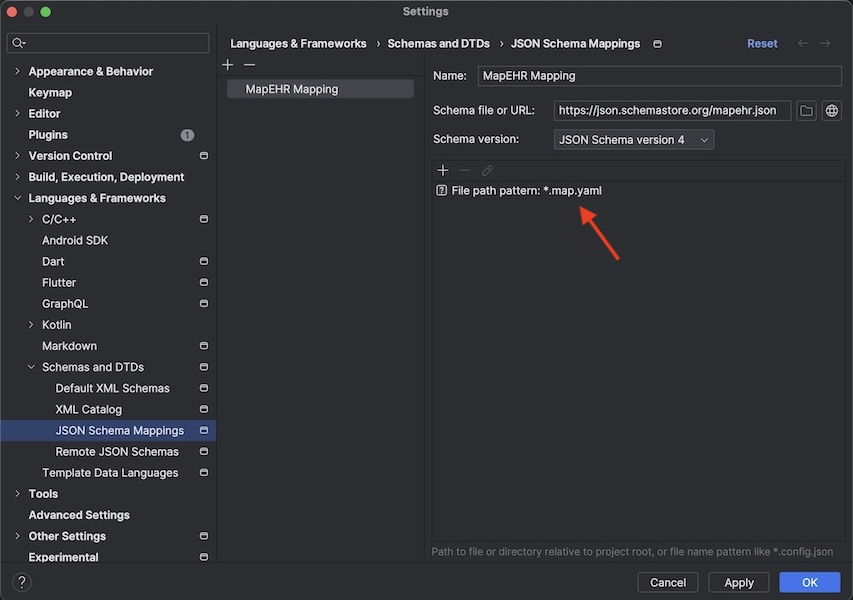
-
Done. Every
.map.yamlfile will not have autocomplete and help in the pop-up window.
For other editors please visit https://www.schemastore.org/json/ and scroll to Supporting editors for instructions.